Additions and corrections
Which similarity measure is better for analyzing protein structures in a molecular dynamics trajectory?
Pilar Cossio, Alessandro Laio and Fabio Pietrucci
Phys. Chem. Chem. Phys., 2011, 13, 10421–10425 (DOI: 10.1039/C0CP02675A). Amendment published 11th July 2011.
In the original article, Fig. 1B and 2B erroneously use a dihedral distance defined as 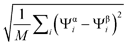 (using the same notation of the article). We present in the following
the same figures done by using the definition for the dihedral distance
(using the same notation of the article). We present in the following
the same figures done by using the definition for the dihedral distance 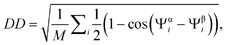 presented on page 10422. The two metrics are extremely well correlated
and coincide for small dihedral differences, as can be checked by Taylor
expanding the second metric. This change does not affect any of the
results or conclusions of the article.
presented on page 10422. The two metrics are extremely well correlated
and coincide for small dihedral differences, as can be checked by Taylor
expanding the second metric. This change does not affect any of the
results or conclusions of the article.
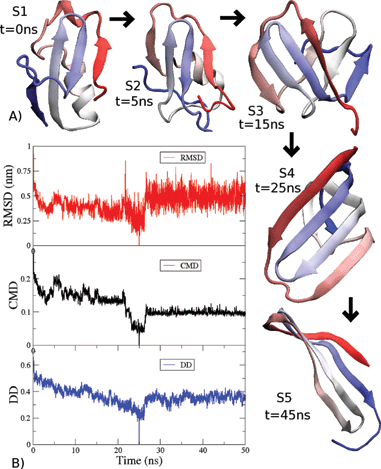
Fig. 1 (A) Different structural conformations observed in a
Val60 trajectory. (B) Similarity measures RMSD, CMD and DD calculated
for all the structures in the trajectory with respect to structure S4.
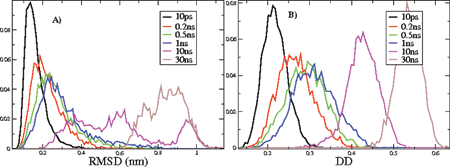
Fig. 2 Probability distributions PΔt of (A) RMSD (nm) and (B) DD for pairs of structures separated by different time intervals Δt.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
Back to article
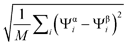 (using the same notation of the article). We present in the following
the same figures done by using the definition for the dihedral distance
(using the same notation of the article). We present in the following
the same figures done by using the definition for the dihedral distance 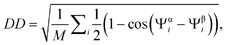 presented on page 10422. The two metrics are extremely well correlated
and coincide for small dihedral differences, as can be checked by Taylor
expanding the second metric. This change does not affect any of the
results or conclusions of the article.
presented on page 10422. The two metrics are extremely well correlated
and coincide for small dihedral differences, as can be checked by Taylor
expanding the second metric. This change does not affect any of the
results or conclusions of the article.
