We report a set of tools to be used in conjunction with a robot-based Internet indexing engine which can be used to convert non-conforming HTML collections to well-formed and valid XHTML documents. The tools, inter alia, can correct invalid syntax which can occur in embedded RasMol scripts and extract chemical meta-information from normally inaccessible document components, including transcluded chemical files. The index that can be built from the transformed documents can be used to improve the quality of searches carried out in a chemical context.
A large amount of chemical and molecular data and information has
accumulated over the last six years in the form of marked up hypertext
documents stored on World Wide Web servers.1 The inter-linked nature
of such documents has, in turn, spawned a generation of robot-based methods
which can traverse the document collection to compile a searchable index.
However, the early adoption of what can now be recognised as immature standards
for authoring the documents has resulted in a number of serious structural
deficiencies in the indexing and searching of such document collections. More
particularly, the web is often regarded as a relatively low quality chemo and
bio-informatics resource discovery tool, because of the difficulty in searching
for and identifying chemical content.
In the present article, we discuss our strategy for creating mechanisms for identifying chemical information in this medium. We introduce the basic design principles for creating web-based documents, and then describe further modifications to the indexing robot described in earlier articles2 which allow the transformation of most existing chemical document collections to a modern form based on XML standards.3 Procedures for identifying or deriving specific chemical content and expressing this as meta-information are established, and we conclude by describing some chemical search interfaces for the resulting indexes.
One aspect of the evolution of hypertext markup language (HTML) during the period 1993–1999 has been greater recognition of the rigour required for the document structure and syntax. Three important principles, in particular, need stating.
(i) The three basic components of a document should be clearly identified and clearly separated. These are: the data or content of the document, its style or presentation to the user and any logic (e.g. inlined Javascripts) used to interpret the content.
(ii) The document itself should be well formed. In a markup language such as HTML or XML, the syntax of the components should be precisely defined in a standard manner, such that each component (termed an element) is clearly separable from the others. Each element must have well-defined attributes, and the values of these attributes must be expressed in syntax which does not conflict with the document syntax.
(iii) The components of the document should be capable of validation. By this we mean that a definition is available of what elements are allowed in the document, and that this definition specifies what attributes of each element are allowed, including any possible limitations or boundaries on the values of the attribute.
The first version of HTML to formally require such adherance is known as XHTML.4 The first phase of our work involved the development of a tool which we termed JChemDig and which was designed to automate, where possible, the conversion and normalisation of non-compliant older forms of HTML to XHTML.2 Our procedure involved extending software based on the HtDig web robot by adding chemical components written in Java. To this was added a set of XHTML conversion tools, again with added chemical functionality, which we termed JChemTidy.
We now describe a second phase in the development of this suite of tools, with two objectives. We address some further limitations of "legacy" HTML by developing additional tools to correct the syntax used to express the value of certain element attributes. Secondly, we use meta-information declarations and links to allow robot-based indexing engines which do not formally implement the advanced features of XHTML to index the document collection more fully. The implementation of solutions for these specific cases is described in the next section.
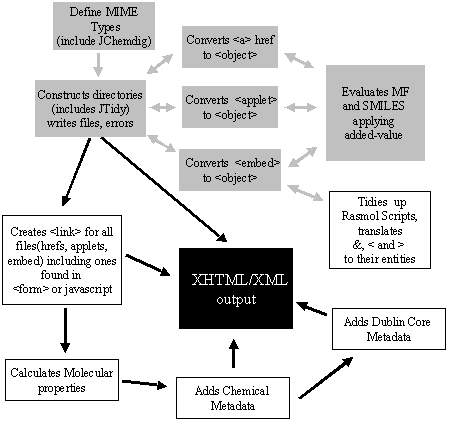
The additional classes added to the previously described JChemTidy set
are indicated in Table 1. A schematic of how these new JChemMeta classes
inter-operate with those previously described is shown in Fig. 1.
Table 1 JChemMeta classes
| JChemMeta class name | Description |
|---|---|
| HtDigfrontmain.class | External parser called from HtDig robot. Calls corresponding classes according to MIME type. |
| HtmlConvert.class | Constructs directories, calls JTidy, writes converted HTML file, reports errors. |
| ScriptTranslator.class | Searches HTML document for scripts in an <embed> tag, translates &, < and > to their entities, adds ; at the end of each script line, formats according to Scheme 1. |
| addMetaData.class | Adds Dublin Core metadata. |
| ExtractHref.class | Searches HTML document for links to other html or chemical files in anchors, <embed>, <applet>, <form> or javascript. Creates new metatags and formats metatag declarations for these files according to Scheme 2. |
| ExecProcess*Href.class (* = Mol, pdb, xyz) | Evaluates SMILES string using external process, creates chemical metadata. |
| read*Href.class (* = Mol, pdb, xyz) | Evaluates molecular formula from contents of local file, creates chemical metadata. |

|
| Fig. 1 Function of JChemMeta classes. Grey boxes represent the functionality reported previously.2 White boxes indicate those classes described in the present article. |
In our development of JChemTidy,2 we focused primarily on use of the <object> element or tag, and particularly on the context of invoking a chemical object.5 This element, which is both well formed and validatable, replaces the older and now deprecated <embed> element, which has been frequently used to insert (transclude) chemical information onto a web page. Use of the <object> element will often require additional element attributes such as "id" or "title" to be added to the original descriptor, information which many index engines do not yet process during the indexing process. Subsequently, we have identified a number of further desirable enhancements to JChemTidy, which are discussed below.
Many invocations of the <embed> element include a script attribute, the value of which is a so-called RasMol script (Scheme 1). These scripts can contain the characters < and >, which are used as atom selection operators, but which when embedded inside a XHTML document will prevent it from being well formed. Such characters need replacing by quoted references, known as entities.
| RasMol script code (incompatible for XHTML conversion) |
Converted RasMol script code (compatible for XHTML conversion) |
XHTML converted code (JChemTidy)2 |
|---|---|---|
<embed border="0" |
<embed border="0" |
<object data="chair.pdb" |
| Scheme 1 |
RasMol scripts comprise a sequence of individual commands which are separated by either a semi-colon or, very often, an end-of-line character (also known as a line break). The latter, in particular, is operating system specific, and its use is not recognised in an XHTML document, which derives its structure from markup rather than line breaks. Since removal of such line breaks would destroy the meaning of the RasMol script, their locations need to be replaced by an appropriate semi-colon character.
It has become increasingly common to transclude chemical content in an HTML document via a <form> element. Such an element is now seen as intermixing data (i.e. a URL specifying a link to another document) with logic (an action resulting from a user selection of various options). Such implicit links to other documents are now formally regarded as better suited for specification via a <link> element, and in the future, via the XFORM standard.6
It has also become common for certain aspects of window handling (their size, position, content, etc.) to be achieved using HTML <script> elements. Thus, the linked content of an HTML document might be specified by the logical components of that document, a form in which it is, in general, very difficult indeed to formally identify the content without following the logic. In certain specific instances however, when, for example, an external chemical document or datafile is transcluded via a declaration of its name, it is possible to automatically respecify this document using a formal <link> element. The general issue of the dynamic creation of document content using e.g. JavaScript logic in combination with user-supplied variables remains a serious one if the formal identification of document components and resources using automatic indexing methods is desired.
An initial pass over a document collection serves simply to normalise any RasMol scripts declared as <embed> element attributes, removing line breaks and angle brackets, < and > (Scheme 1). This will allow each document to be presented to the JTidy HTML to XHTML conversion tool in such a manner which will not cause the document to be rejected by JTidy.
During a second pass, we add the capability to recognise references to chemical or HTML documents as components of a <form> element and these are automatically added to the derived XHTML version of the document via a <link> declaration, using relative rather than absolute paths for the URL (Scheme 1). The link type is derived from the chemical MIME type of the referenced chemical datafile and is inserted as a child of the document <head> element.
At this stage, all <embed> and <applet> elements are replaced by <object> declarations. To each object, a title attribute which contains a derived declaration of its content is added. For example, if it relates to a chemical datafile, a string of the type title="Molecule-object, Molfile-format, SMILES: CO[BH2]OC=C" is inserted. The addition of titles to such elements allows the chemical components to be clearly identified in subsequent indexing and searching of the document.
The <head> section of an XHTML document can also contain the <meta> element, comprising meta-information about the important components of the document. Such meta-information is used by indexing engines to impart greater significance to the weighting of any individual document in a collection. It also serves to provide a standard location and format for key information that might otherwise be located in less accessible parts of the body of the document, for instance, in <script> or <form> elements, as noted above. Because meta-declarations are frequently omitted from authored HTML documents, or worse, are inherited as templates from other documents, we felt it desirable to implement some degree of meta-information handling in our procedure. Dublin Core metadata can be automtically added to a document by opening a URL connection with the Dublin Core Generator.7 The output of this process takes the form of a well-formed document, and it is trivial to extract the meta elements and add them to our XHTML version of the document. We also have the option of adding meta-information8 in the form of derived values for any chemical files transcluded as <embed> or <object> elements to the document <head>. These could include, for example, a molecular formula resulting from analysis of e.g. a Molfile, or a unique SMILES string derived from passing the Molfile to an external program. Although we illustrate only two such <meta> fields here (Scheme 2), in principle, any external process can be used to add such information. We have used both the standard Dublin Core schema and an extended chemical schema defined by us.8
| Original HTML code | XHTML code with derived metadata and links |
|---|---|
<a href="cGMP.mol">anchor</a>
<form name="GetMolecule">
<option value="cGMP.mol"
selected="selected">
</option>
</form>
<script language=javascript>
function presentMolecule()
{window.open('cGMP.mol','JME','
width=680,height=530');}
</script>
<embed src="cGMP.mol" width="250" height="250"
name="chair" script="script.spt">
<object data="cGMP.mol" width="250" height="250">
<param name="name" value="chair"/>
<param name="script" value="script.spt"/>
.
.
<object>
|
<!-- Links extracted from Form and Script --> <link type="chemical/x-mdl-molfile" rel="alt" href="cGMP.mol" title="This is a link to a MDL Molfile"/> <!-- Links extracted for RasMol Scripts --> <link type="application/x-spt" rel="alt" href="script.spt" title="This is a RasMol script file"/> <!-- Derived meta-information for cGMP.mol --> <!-- Dublin Core Schema --> <meta name="DC.Subject" content="Molecule-href, Molfile-format"/> <!-- Chemistry schema --> <meta name="DC.chem.substance.formula" scheme="formula" content="C10H11N5O7P1"/> <meta name="DC.chem.substance.smiles" scheme="smiles" content="Nc1nc2n(cnc2c(=O)[nH]1)C3OC(CO)C4OP(=O)([O-])OC34"/> <link rel="DC.chem.substance.smiles" type="text/html" href="http://www.daylight.com/dayhtml/doc/theory/theory.smiles.html"> <!-- Derived meta-information for RasMol Scripts --> <meta name="DC.Subject" content="RasMol-script"/> |
| Scheme 2 |
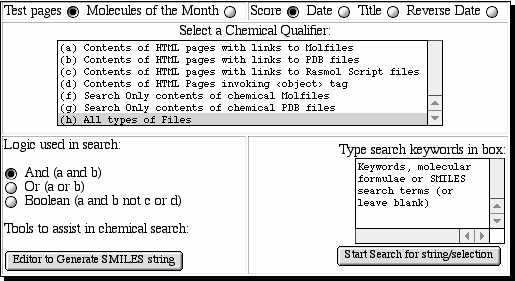
Few current search engines will follow the content of <embed>, <applet> or <object> tags and none that we know of will follow the <form> or <script> elements. By adding the appropriate links to the <head> component of our derived XHTML version of the document, we can enable any descent search engine to follow such content. To illustrate a typical search, we have used the HtDig system itself to re-index the derived XHTML document collection. This enables searches to be qualified with chemically coarse-grained restrictions (Fig. 2). For example, one can limit the search to only those HTML documents which transclude an MDL Molfile [qualifcation (a)], a PDF format file [qualification (b)] or, more generally, any declared <object> [qualification (c)]. Further examples include documents invoking RasMol scripts, either internally or by referencing an external script [qualifcation (d)]. Other types of transcluded file can, of course, also be included in the indexing operation and subsequent search.
Boolean logic (AND, OR) can be used in conjunction with additional specified keywords. These keywords can be conventional bibliographic search terms, or they can include molecular formulae or derived values, such as SMILES molecular descriptors for molecules transcluded via Molfiles or PDB files. We note in this context that the same canonicalising algorithm must be used in deriving the SMILES search string as in creating it for the index. For example, we include a button to invoke the JME structure editor, which can perform both functions. We emphasize that this approach cannot be used for sub-structure searching, for example. The Boolean operations can also be extended to searching for all HTML documents which include a link to e.g. both Molfile AND PDB files by making a multiple selection (normally using the shift key) from the menu.
Finally, we have implemented an option for searching directly for the content of the Molfile and PDB file itself, rather than the document that invokes it. This makes use of the external parser that we have developed as part of the ChemDig tools for such files. These parsers index only the bibliographic content of such files, typically author, comment, title or remark fields contained within the file. In principle, the RasMol script references could also be indexed and searched, although we have not yet implemented this option.
|
| Fig. 2 Chemically qualified searches. |
The net result of these overall operations is the production of a set of well-formed, valid XHTML documents in which discrete chemical components derived from chemical MIME type datasets are identified with appropriate header information, as well as specified via <object> elements. The use of meta-information is also designed to facilitate linking across information disciplines. We have achieved a limited degree of separation of the logical components of the original document (<form> and <script>) from the data component. A general solution to the problem of formal separation of content from the logic of a document remains to be achieved. One particular issue which we note here is the desirability of indexing RasMol scripts. These can be regarded as the molecular equivalent of the <style> element in XHTML, but can also carry a wealth of information about the molecular coordinate files, either in the form of in-line comments, or in relation to operations on components or regions of the molecule.
The approach described here is suited to indexing coarse-grained chemical content, in which the existence of molecular components, such as atom coordinates, spectra, etc., can be identified, together with some limited derived information, such as a molecular formula or atom connection descriptors. Achieving finer granularity of chemical information will require more precise document markup. In this context, we note that the XHTML documents that our tools can be used to create are well suited for further transformations to finely grained XML components using e.g. chemical markup language (CML).3 Taken with XML-based resource description frameworks such as RDF,9 the move towards the "semantic web" envisioned by its principle architect comes one step closer.
G. V. G. thanks Merck Sharp and Dohme and the EPSRC for the award of a
studentship.
This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2001