Enhancement of the Chemical Semantic Web through InChIfication
Simon J. Colesa, Nick E Dayb, Peter Murray-Rustb, Henry S. Rzepac and Yong Zhangb.
aSchool of Chemistry, University of Southampton, Southampton, SO17 1BJ, UK bUnilever Centre for Molecular Informatics, Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge. CB2 1EW, UK cDepartment of Chemistry, Imperial College, London, SW7 2AY, England.
Molecules, as defined by connectivity specified via the IUPAC International Chemical Identifier (InChI), are precisely indexed by major web search engines so that Internet tools can be transparently used for unique structure searches.
Search Engines and strategy
To examine the ability of today's search engines at indexing and returning both CAS numbers and InChIs, searches were performed on:
Search engines used:
| Name | URL |
| GoogleTM | http://www.google.com |
| AOL SearchTM | http://search.aol.com |
| YahooTM | http://www.yahoo.com |
| AltavistaTM | http://www.altavista.com |
| MSN SearchTM | http://search.msn.com |
| Ask JeevesTM | http://www.ask.com |
| TeomaTM | http://www.teoma.com |
| DogpileTM | http://www.dogpile.com |
This covers the three most popular search engine 'providers' (of free listings) in GoogleTM, YahooTM and TeomaTM and also others for which they provide main listings (GoogleTM <-- AOL SearchTM, YahooTM <-- AltavistaTM/MSN SearchTM, TeomaTM <-- Ask JeevesTM). Also included was DogpileTM, a popular Meta search engine that draws results from a number of other search engines1 (including GoogleTM, YahooTM, Ask JeevesTM and OvertureTM) and returns the ones it considers relevant. We believe that engines in different countries may give different results but this will probably not affect our subject matter and this was not controlled for.
It is extremely important to realise that the analysis of search engines is an inexact science. Search engines do not describe their indexing and retrieval methods (presumably to protect competitive advantages) and may change their strategies at frequent intervals to deter manipulations of rankings. The number of indexed pages changes every minute and a search is, ipso facto, not reproducible. We therefore quote approximate times that searches were performed and many of our conclusions should be adjusted for this. However the results we show have such clear outcomes that we are confident that variations in time and place do not affect them. In the following discussion all searches are in bold type.
It is not clear which pages are indexed by search engines but we believe the following:
- Only static web pages are indexed. Pages created on-the-fly as a result of database queries seem to be ignored.
- All engines seem to index X/HTML, DOC, PDF, TXT but may not always index XML (or CML) files. Searching is performed on the ASCII content (XML markup and formatting is ignored).
- All pages fitting the selection above are indexed; no documents are rejected because they are meaningless to humans. There is no reason why any chemical documents should not be included.
By default most engines appear to use the following strategy:
- Use case-insensitive strings, we believe this cannot be altered. This is unlikely to cause many problems in practice; although, for example, CO and Co would collide there is normally enough syntactic context to remove ambiguity.
- Do not carry out stemming and other lexical operations in the presumed language of the query. Thus GoogleTM-2004-11-21 retrieves 77 documents for datument and 111 for datuments. There is apparently no commonality (unless the documents contain both words).
-
Use a list of stop words. If GoogleTM-2004-11
is given "to be or not to be, that is the
question". It returns:
The following words are very common and were not included in your search: to be to be that is the. Lowercase "or" was ignored. Try "OR" to search for either of two terms
and searches only for not and question. This gives many false positives. - By default search engines now seem to apply AND operations between multiple terms so that wallaby datument returns no hits on any engines (2004-11-21). This appears to be a change in policy from earlier versions which allowed some OR results to be returned with low numbers of hits.
- Strings can be found anywhere in the document. Thus chemical semantic web returns ca. 70, 000 hits, most of which contain the three tokens in widely separated positions. We believe, however, that some engines will order results to emphasize proximity.
-
Tokenize the query at any punctuation, Thus
AltavistaTM-2004-11-21 returned 298 results
for murray-rust wallaby. A typical one included
the summary:
... Dance of Death (album) David Murray. Enhanced CD ... British Heavy Metal. Power metal. Rust. Slayer ... banded gecko. Banded Hare-Wallaby (enc.) ...
Note that the search terms are not required to be proximal and can be de-hyphenated or re-hyphenated. -
Hide or coalesce "similar" results. Thus
GoogleTM-2004-11-21 returns 3 results for
"114795-97-0" but adds the message:
In order to show you the most relevant results, we have omitted some entries very similar to the 3 already displayed.
Following this link now gives a total of 4 hits. This strategy was essential in analysing the recall and precision of InChIs, as different InChIs can appear "similar" to search engines.
If you like, you can repeat the search with the omitted results included.
All search engines appear to allow refinements of the query to increase precision, and seem to use a common syntax and behaviour. Some options can be provided by syntax but most are on a special "Advanced Search" page.
- Tokens can be mandatory (prefixed by +) or disallowed (-). Thus +chemical +semantic +web would recall only those pages which contain all three words. However search engines frequently seem to adopt this strategy by default and the OR option needs to be switched on.
- Enclosing strings in quotes requires that all tokens be returned in the precise order and juxtaposed. Thus semantic chemical web would recall chemical semantic web but "semantic chemical web" would not. However any punctuation appears to be normalised to whitespace, so that "(chemical) semantic-web" returns the same hits as"chemical semantic web".
-
Strings can be long; we have not ascertained a maximum
length but GoogleTM-2004-11-21 retrieves a
60-character protein sequence fragment such as
EPTTMITLGPLLVFLVIGFFAWLLFTLAVFALPVFAGVTIGLWAFHTGAGALGGIAVGPV
while a substring, as expected, is not retrieved.
Search engines seem to have a maximum number of tokens in a quoted string; apparently 10* in GoogleTM-2004-11 and AltaVistaTM-2004-11. A GoogleTM-2004-11-21 search for "twas brillig and the slithy toves did gyre and gimble in the wabe" returns 4020 hits and advises "in"(and any subsequent words) was ignored because we limit queries to 10 words. So the same hits are returned for the string "twas brillig and the slithy toves did gyre and gimble in the bath". Initially this is counted as false positive, but it is easily eliminated by a textual search of the document. This means that a simple tool can match the full text of the retrieved documents and eliminate hits that did not contain the full search string. This is important for InChIs which almost always have more than 10 tokens.
Search engines also seem to have a limit to the number of the recalled entries you can view. On searching for a common word you may be informed that there are 10,000 documents found that match, but you will never be able to view them all. Below is a table with the maximum number of results that can be viewed for each search engine (2004-11-21):
| GoogleTM | AOL SearchTM | YahooTM | AltavistaTM | MSN SearchTM | Ask JeevesTM | TeomaTM | DogpileTM | |
| No. of viewable entries | ~1000 | 535 | 1000 | 1050 | 1000 | 200 | 200 | ~125 |
This could prove a problem for the future, particularly if someone were to search for a large molecule which had more stereoisomers than viewable results from the search engine.
Search terms and metrics
In information retrieval (IR) it is normal practice to define a precise corpus which is to be searched (or processed) and to measure the recall and precision of different strategies. In this case the unit of measurement is the page (or document). It is possible that a page may contain multiple instances of search terms but this was not relevant here. We use the abbreviations and terms:
- P = total known pages
- H = total pages retrieved (hits)
- TP = true positive; all those retrieved pages which match the InChI
- FP1 = false positive matching another InChI
- FP2 = false positive matching a non-InChI string
- FP = total false positive
- FN = false negative; pages containing the query InChI which were not retrieved
FP = FP1 + FP2
H = TP + FP
P = TP + FN
recall = TP / P
precision = TP / H
These concepts are not applicable when the size of the corpus is unknown, as for the searches for CAS registry numbers.
Searching for CAS numbers
Chemical Abstracts registry numbers ("CAS numbers") are widely used across the World Wide Web and can also act as unique identifiers. To test precision and recall two very common compounds, caffeine and acetic acid were chosen. These occur in many types of document (journal articles, suppliers' catalogs, Materials Safety Data Sheets, lists of properties) and in multiple sources. The precise number of web pages containing a given CAS number is unknown and changes continuously, so that recall cannot be established. (As CAS numbers are copyright, we assumed it may not be legal to create test documents without permission). In practice authors of web pages use a variety of syntaxes such as CAS: 64-19-7, CAS number: 64-19-7 , Registry number: 64-19-7 and frequently simply 64-19-7 (almost universal when tables are used). Recall will obviously be higher for the pure number but precision will be lower.
It is impossible to measure the precision and recall of the whole corpus (often > 10^4 entries). The first 100 hits with 3 strategies were therefore analysed. For each strategy the format is:
True positives/sample size
(Total hits)
Table 1: Searching for CAS numbers with various strings (2004-11-18)
Acetic acid: 64-19-7
| GoogleTM | AOL SearchTM | YahooTM | AltavistaTM | MSN SearchTM | Ask JeevesTM | TeomaTM | DogpileTM | |
| "64-19-7" | 79/100 (15,100) |
78/100 (3220) |
9/100 (10,200) |
27/100 (9,830) |
63/100 (1,486) |
82/100 (2,950) |
82/100 (2,950) |
28/48 (48) |
| +CAS +"64-19-7" | 100/100 (10,800) |
99/100 (2,320) |
99/100 (4,400) |
99/100 (4,250) |
100/100 (881) |
100/100 (1,530) |
100/100 (1,530) |
62/63 (63) |
| +CAS +number +"64-19-7" | 100/100 (5,300) |
100/100 (1125) |
100/100 (1,380) |
100/100 (1,500) |
99/100 (495) |
100/100 (1,060) |
100/100 (1,060) |
60/61 (61) |
| Caffeine: 58-08-2 | ||||||||
| "58-08-2" | 28/100 (6720) |
28/100 (1435) |
0/100 (543,000) |
0/100 (550,000) |
0/100 (100,488) |
43/100 (2,540) |
43/100 (2,540) |
9/11 (11) |
| +CAS +"58-08-2" | 100/100 (873) |
100/100 (550) |
23/100 (13,300) |
20/100 (13,400) |
21/100 (2,289) |
98/100 (396) |
98/100 (396) |
21/22 (22) |
| +CAS +"number" +"58-08-2" | 100/100 (8,250) |
100/100 (265) |
32/100 (4,430) |
34/100 (2,080) |
25/100 (601) |
94/100 (207) |
94/100 (207) |
30/56 (56) |
Note that the total number of hits varies enormously and that the aggregator (DogpileTM) is clearly selective. MSN SearchTM seems to select on single tokens and neglects order. The contrast in precision between caffeine and acetic acid (YahooTM, MSN SearchTM, TeomaTM) is surprising since the actual tokens are probably relatively equifrequent. Some differences may be due in part to speed of indexing. It is interesting that adding the apparent constraint +number to the search actually increases the total hits.
Manual examination of the first 100 entries showed the number of false positives (i.e. not chemical compounds). Not surprisingly this can be a high percentage for the raw strings as they retrieve many other triads (dates, phone numbers, etc.) and the enormous amount of noise for 58-08-02 seems to be due in part to ringtones. It seems that the string CAS provides complete recall in some cases but at the expense of precision (< 50%).
Searching for InChIs
The InChI architecture and implications
An InChI string consists of layers, the first being the chemical formula of the compound. This is followed by the atom connection information, which in turn is followed by optional layers containing information such as stereochemistry or isotopic content. For most molecules there are many more than 10* tokens that appear before the connection information is complete. We can see that when searching for a stereoisomer of a large molecule, that none of the information in the stereochemical layer of the InChI string will be included in a search by GoogleTM. Thus if InChI strings of other stereoisomers were on the web, then they would be seen as identical to the search string and incorrectly returned. The same can be said of any molecules with the same connection information but differing information in the later layers. Indeed, it would also be possible for two different molecules to have the same chemical formula and also start of the connection information but have differing connection information in the regions not searched for by GoogleTM. So unless search engines start searching with the whole search string entered, this could cause minor problems in the future. If not, a program used post-search to scan the recalled entries for the complete InChI string would be necessary to ensure no 'other-InChI' false positives.
Results
There are very few InChIs on the web, so our experiment was performed on a bounded dataset of unique compounds. We chose the University of Southampton's Crystal Structure Report Archive website2, which at the time (2004-11-18) contained 104 pages each containing the results of a crystal structure. Each page contains an IChI (sic) string, created with version 0.932Beta of the identifier. Note that the compunds are mainly novel and/or complex so it is extremely unlikely that anyone outside the authors will have published the same compounds using IChIs in a different context, especially as V0.932 beta is now obsolete. We therefore have an accurate estimate of recall as totalHits / totalKnownPages.
This corpus consists of:
- 104 HTML pages
- 100 describing different molecules for which the IChI is unique
- 2 pages describing different experiments on the same compound and will therefore have the same IChI
- 2 pages describing a pair of diastereomers which have the same IChI as stereochemistry was omitted from its calculation. This is an inter-IChI collision which the search engines obviously cannot detect.
- 93 CML pages which are mapped to the HTML pages
- 89 different molecules with unique IChIs
- 2 of the two experiments on the same molecule
- 2 of the two diastereomers
There are thus a total of 91 IChIs on the 93 Southampton CML pages.
To simplify the analysis we report the HTML and CML retrieval separately.
For each of the 102 IChIs a separate search was performed on each engine, using the quoted IChI string. The results are aggregated in Table 2 and a typical resolved item is shown in Figure 1
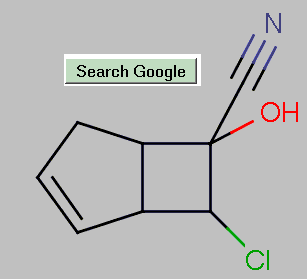 |
 |
Table 2: Recall of InChI strings from the Crystal Structure Report Archive2 (2004-11-18)
The results of the search for each engine are aggregated within cells (described in the caption). The search was performed on two dates and shows a significant increase in the MSN recall.
TP / FP1 / FP2 / P / recall (%) / precision (%)
| Pages | GoogleTM | AOL SearchTM | YahooTM | AltavistaTM | MSN SearchTM | Ask JeevesTM | TeomaTM | DogpileTM | |
| 2004-11-18 | |||||||||
| .html | 104 | 104/0/0/100/100 | 102/0/0/98/100 | 33/0/0/32/100 | 39/0/0/38/100 | 43/0/0/42/100 | 0/0/0/0/- | 0/0/0/0/- | 102/0/0/98/100 |
| .cml | 93 | 92/0/0/99/100 | 91/0/0/98/100 | 0/0/0/0/- | 0/0/0/0/- | 0/0/0/0/- | 0/0/0/0/- | 0/0/0/0/- | 91/0/0/98/100 |
| 2004-11-05 | |||||||||
| .html | 104 | 103/0/0/99/100 | 15/0/0/14/100 | 20/0/0/19/100 | 0/0/0/0/- | ||||
| .cml | 93 | 67/0/0/72/100 | 0/0/0/0/- | 0/0/0/0/- | 0/0/0/0/- | ||||
Thus out of 832 searches performed on 8 different search engines there were no false positives.
Why is there a difference in recall between InChI strings and CAS numbers?
A CAS number consists of only numbers separated by generic punctuation and an InChI string consists of blocks of letters and numbers separated by generic punctuation. CAS numbers are at a disadvantage as they are short and only contain numbers and generic punctuation. Indeed, when searching for the CAS number of caffeine 58-08-2 it is quite common for documents containing information on acetone to be recalled, as its molecular weight is 58.08. InChI strings are generally much longer than CAS numbers and as they have a good mix of letters and numbers in their tokens and separation by generic punctuation, it is unlikely that even a small section of an InChI string will be matched to anything else on the World Wide Web.
Searching for SMILES
In principle SMILES is uniquefiable [3] but in practice there is no public conformance to the specification and we also believe that some implementations produce incompatible results. The present study confirms the variation. To determine the usage of SMILES, queries of the form SMILES AND ("caffeine" OR "58-08-2") were used. This is clearly imprecise, but about 10 sites were found containing SMILES for caffeine which showed at least 7 different syntactic variants. To test precision and recall these were submitted to GoogleTM-2004-11-20.
True positives/False
positives/non-SMILES
(Total hits)
| Search String | GoogleTM | Located on Sites |
| "[c]1([n+]([CH3])[c]([c]2([c]([n+]1[CH3])[n][cH][n+]2[CH3]))[O-])[O-]" | 2/3/0 (5) |
www.biocheminfo.org www.eureka.ya.com www.biozentrum.unibas.ch |
| "CN1C(=O)N(C)C(=O)C(N(C)C=N2)=C12" | 14/0/0 (14) |
www.daylight.com |
| "Cn1cnc2n(C)c(=O)n(C)c(=O)c12" | 20/0/0 (20) |
www.daylight.com pubs.acs.org www.predictive-toxicology.org www.eyesopen.com bind.ca www.surrey.ac.uk www.sunsetmolecular.com |
| "Cn1cnc2c1c(=O)n(C)c(=O)n2C" | 2/0/0 (2) |
www.molinspiration.com doi.wiley.com |
| "N1(C)C(=O)N(C)C2=C(C1=O)N(C)C=N2" | 1/0/0 (1) |
www.fda.gov |
| "O=C1C2=C(N=CN2C)N(C(=O)N1C)C" | 2/0/0 {2} |
potency.berkeley.edu |
| "CN1C=NC2=C1C(=O)N(C)C(=O)N2C" | 17/0/0 (17) |
www.jchem.com www.chemaxon.com www.structuresearch.com www.chemaxon.hu bohlmann.bgbm.org |
The raw precision of the above SMILES strings is high. This can be attributed to each string (apart from the first) having around 10* tokens, allowing Google to search for the whole string and match it exactly with strings found on the web. The first string has 18 tokens, and so a large part is not included in the search, leading to lower precision. To a search engine SMILES strings are similar to InChI strings as they consist of tokens containing letters and numbers separated by generic punctuation.
The 58 pages occurred on 20 sites with 7 syntactic variants. There is thus no commonality of approach (i.e. the page creators are not using a synoptic approach).
Searching for InChI strings from the KEGG Collection using Google
On 2004-10-04 9585 molecules from the KEGG collection were converted to CML, indexed with InChI V1.12 Beta and posted as static pages on the WWW. To our knowledge there are very few other InChI V1.12 Beta instances on the web, so this provides a test of recall.
At 2004-11-16 the molecules at wwmm.ch.cam.ac.uk/data/kegg have been indexed on GoogleTM up to c07576. As far as we know the indexing is serial so that 4870 molecules were indexed. To test precision and recall the InChIs for 83 KEGG ligands c00001-c00100 were submitted to GoogleTM. (Table 3)
There were no non-InChI recalls (FP1 = 0) and no false negatives (FN = 0). No hits were found to molecules not on our site so recall is measured with respect to this. The FP2 are due to collisions after the first 10* unique tokens. They are easily removed by a simple program filtering the search engine results. For each false positive its quoted InChI was submitted to see if all the recalls were symmetric (i.e. if molecule c00010 recalls c00298, does c00298 recall c00010). There are 35 isomeric collisions in about 4500 molecules, which suggests that the false collisions can be managed with simple filters. In practice, also, many molecules in KEGG do not have complete stereochemistry so that methods other than the connection table (e.g. names) would have to be used to separate them.
Isomers with InChI collisions
Each of the 35 entries contains two of more colliding InChIs, for each of which the serial number (not the KEGG id) and the InChI are given. The entry is optionally followed by a note. Note that all InChI collisions are trivially resolvable after retrieval and are listed to give an idea of the length of the strings that most search engines index. The only problem arises if collisions are so frequent that the search engine cuts off before returning all the true positives.
c01328= 1.12Beta/H2O/h1H2/p-1 c00001= 1.12Beta/H2O/h1H2 NOTE: 1 (water) recalls 1328 BUT 1328 does not recall 1 c00704= 1.12Beta/O2/C1-2 c00007= 1.12Beta/O2/C1-2 NOTE: 704 is the superoxide ion O2- and should have been rendered as 1.12Beta/O2/C1-2/q-1 c00054= 1.12Beta/C10H15N5O10P2/C11-8-5-9(13-2-12-8)15(3-14-5)10-6(16)7(25-27(20,21)22)4(24-10)1-23-26(17,18)19/h1H2,2-4H,6-7H,10H,16H,(H2,11,12,13)(H2,17,18,19)(H2,20,21,22)/t4-,6-,7-,10-/m1/s1 c00008= 1.12Beta/C10H15N5O10P2/C11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(24-10)1-23-27(21,22)25-26(18,19)20/h1H2,2-4H,6-7H,10H,16-17H,(H,21,22)(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 c03850= 1.12Beta/C10H15N5O10P2/C11-8-5-9(13-2-12-8)15(3-14-5)10-7(25-27(20,21)22)6(16)4(24-10)1-23-26(17,18)19/h1H2,2-4H,6-7H,10H,16H,(H2,11,12,13)(H2,17,18,19)(H2,20,21,22)/t4-,6-,7-,10-/m1/s1 NOTE: These are isomers but the connection table only differs after the 10th token c00014= 1.12Beta/H3N/h1H3 c01342= 1.12Beta/H3N/h1H3/p+1 NOTE: 14 recalls 1342 BUT 1342 does not recall 14 c01367= 1.12Beta/C10H14N5O7P/C11-8-5-9(13-2-12-8)15(3-14-5)10-6(17)7(4(1-16)21-10)22-23(18,19)20/h1H2,2-4H,6-7H,10H,16-17H,(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 c04378= 1.12Beta/C10H14N5O7P/C11-8-5-9(13-2-12-8)14-3-15(5)10-7(17)6(16)4(22-10)1-21-23(18,19)20/h1H2,2-4H,6-7H,10H,16-17H,(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 c00946= 1.12Beta/C10H14N5O7P/C11-8-5-9(13-2-12-8)15(3-14-5)10-7(22-23(18,19)20)6(17)4(1-16)21-10/h1H2,2-4H,6-7H,10H,16-17H,(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 c00020= 1.12Beta/C10H14N5O7P/C11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(22-10)1-21-23(18,19)20/h1H2,2-4H,6-7H,10H,16-17H,(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 c00023= 1.12Beta/Fe c00824= 1.12Beta/Fe.H2S/h;1H2/q+1;/p-1 NOTE: 23 recalls 824 BUT 824 does not recall 23 NOTE: c00023 is junk as no proper charge is given c00217= 1.12Beta/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h1-2H2,3H,6H2,(H,7,8)(H,9,10)/t3-/m1/s1 c00302= 1.12Beta/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h1-2H2,3H,6H2,(H,7,8)(H,9,10) c00025= 1.12Beta/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h1-2H2,3H,6H2,(H,7,8)(H,9,10)/t3-/m0/s1 c00029= 1.12Beta/C15H24N2O17P2/C18-3-5-8(20)10(22)12(24)14(32-5)33-36(28,29)34-35(26,27)30-4-6-9(21)11(23)13(31-6)17-2-1-7(19)16-15(17)25/h1-2H,3-4H2,5-6H,8-14H,18H,20-24H,(H,26,27)(H,28,29)(H,16,19,25)/t5-,6-,8+,9-,10+,11-,12+,13-,14?/m1/s1 c00052= 1.12Beta/C15H24N2O17P2/C18-3-5-8(20)10(22)12(24)14(32-5)33-36(28,29)34-35(26,27)30-4-6-9(21)11(23)13(31-6)17-2-1-7(19)16-15(17)25/h1-2H,3-4H2,5-6H,8-14H,18H,20-24H,(H,26,27)(H,28,29)(H,16,19,25)/t5-,6-,8+,9-,10-,11-,12-,13?,14?/m1/s1 c00936= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2?,3?,4-,5-,6-/m0/s1 c00124= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4-,5+,6?/m0/s1 c00159= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4-,5-,6?/m0/s1 c00031= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4-,5+,6?/m0/s1 c06467= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4-,5+,6?/m1/s1 c06464= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4+,5+,6?/m1/s1 c01487= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4+,5-,6?/m1/s1 c00221= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2?,3?,4-,5+,6+/m0/s1 c00267= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4-,5+,6-/m0/s1 c00962= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2?,3?,4-,5+,6+/m0/s1 c00984= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4-,5+,6-/m0/s1 c06465= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4+,5-,6?/m1/s1 c06466= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4+,5+,6?/m1/s1 c00293= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4+,5-,6?/m1/s1 c01582= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4+,5-,6+/m1/s1 c01825= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3+,4+,5-,6+/m0/s1 c02209= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H/t2-,3-,4-,5-,6+/m0/s1 c00738= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H c01381= 1.12Beta/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h1H2,2-11H NOTE: these hexopyranosides (stereoisomers of glucose) show the variation in sterochemical information in KEGG:
- c02209: exact at all centres
- c00738: no stereochemical information given
- c00936: stereochemistry given at 3/5 centres
c02606= 1.12Beta/C4H4O5/c5-2(4(8)9)1-3(6)7/h1H,5H,(H,6,7)(H,8,9)/b2-1- c03981= 1.12Beta/C4H4O5/c5-2(4(8)9)1-3(6)7/h1H,5H,(H,6,7)(H,8,9)/b2-1- c00036= 1.12Beta/C4H4O5/c5-2(4(8)9)1-3(6)7/h1H2,(H,6,7)(H,8,9) c00133= 1.12Beta/C3H7NO2/C1-2(4)3(5)6/h1H3,2H,4H2,(H,5,6)/t2-/m1/s1 c01401= 1.12Beta/C3H7NO2/C1-2(4)3(5)6/h1H3,2H,4H2,(H,5,6) c00041= 1.12Beta/C3H7NO2/C1-2(4)3(5)6/h1H3,2H,4H2,(H,5,6)/t2-/m0/s1 c00203= 1.12Beta/C17H27N3O17P2/C1-6(22)18-10-13(26)11(24)7(4-21)35-16(10)36-39(31,32)37-38(29,30)33-5-8-12(25)14(27)15(34-8)20-3-2-9(23)19-17(20)28/h1H3,2-3H,4-5H2,7-8H,10-16H,21H,24-27H,(H,18,22)(H,29,30)(H,31,32)(H,19,23,28)/t7-,8-,10-,11+,12-,13-,14-,15?,16?/m1/s1 c01170= 1.12Beta/C17H27N3O17P2/C1-6(22)18-10-13(26)11(24)7(4-21)35-16(10)36-39(31,32)37-38(29,30)33-5-8-12(25)14(27)15(34-8)20-3-2-9(23)19-17(20)28/h1H3,2-3H,4-5H2,7-8H,10-16H,21H,24-27H,(H,18,22)(H,29,30)(H,31,32)(H,19,23,28)/t7-,8-,10-,11+,12-,13-,14-,15-,16?/m1/s1 c00043= 1.12Beta/C17H27N3O17P2/C1-6(22)18-10-13(26)11(24)7(4-21)35-16(10)36-39(31,32)37-38(29,30)33-5-8-12(25)14(27)15(34-8)20-3-2-9(23)19-17(20)28/h1H3,2-3H,4-5H2,7-8H,10-16H,21H,24-27H,(H,18,22)(H,29,30)(H,31,32)(H,19,23,28)/t7-,8-,10-,11-,12-,13+,14-,15-,16?/m1/s1 c00047= 1.12Beta/C6H14N2O2/c7-4-2-1-3-5(8)6(9)10/h1-4H2,5H,7-8H2,(H,9,10)/t5-/m0/s1 c00739= 1.12Beta/C6H14N2O2/c7-4-2-1-3-5(8)6(9)10/h1-4H2,5H,7-8H2,(H,9,10)/t5-/m1/s1 c00049= 1.12Beta/C4H7NO4/c5-2(4(8)9)1-3(6)7/h1H2,2H,5H2,(H,6,7)(H,8,9)/t2-/m1/s1 c00402= 1.12Beta/C4H7NO4/c5-2(4(8)9)1-3(6)7/h1H2,2H,5H2,(H,6,7)(H,8,9)/t2-/m1/s1 c00055= 1.12Beta/C9H14N3O8P/C10-5-1-2-12(9(15)11-5)8-7(14)6(13)4(20-8)3-19-21(16,17)18/h1-2H,3H2,4H,6-8H,13-14H,(H2,10,11,15)(H2,16,17,18)/t4-,6-,7-,8-/m1/s1 c05822= 1.12Beta/C9H14N3O8P/C10-5-1-2-12(9(15)11-5)8-6(14)7(4(3-13)19-8)20-21(16,17)18/h1-2H,3H2,4H,6-8H,13-14H,(H2,10,11,15)(H2,16,17,18)/t4-,6-,7-,8-/m1/s1 c03104= 1.12Beta/C9H14N3O8P/C10-5-1-2-12(9(15)11-5)8-7(20-21(16,17)18)6(14)4(3-13)19-8/h1-2H,3H2,4H,6-8H,13-14H,(H2,10,11,15)(H2,16,17,18)/t4-,6-,7-,8-/m1/s1 c00062= 1.12Beta/C6H15N4O2/c7-4(5(11)12)2-1-3-10-6(8)9/h1-3H2,4H,7-9H2,10H,(H,11,12)/t4-/m0/s1 c00792= 1.12Beta/C6H15N4O2/c7-4(5(11)12)2-1-3-10-6(8)9/h1-3H2,4H,7-9H2,10H,(H,11,12)/t4-/m1/s1 c02385= 1.12Beta/C6H15N4O2/c7-4(5(11)12)2-1-3-10-6(8)9/h1-3H2,4H,7-9H2,10H,(H,11,12) c00064= 1.12Beta/C5H10N2O3/c6-3(5(9)10)1-2-4(7)8/h1-2H2,3H,6H2,(H2,7,8)(H,9,10)/t3-/m0/s1 c00819= 1.12Beta/C5H10N2O3/c6-3(5(9)10)1-2-4(7)8/h1-2H2,3H,6H2,(H2,7,8)(H,9,10)/t3-/m1/s1 c00303= 1.12Beta/C5H10N2O3/c6-3(5(9)10)1-2-4(7)8/h1-2H2,3H,6H2,(H2,7,8)(H,9,10) c00716= 1.12Beta/C3H7NO3/c4-2(1-5)3(6)7/h1H2,2H,4H2,5H,(H,6,7) c00065= 1.12Beta/C3H7NO3/c4-2(1-5)3(6)7/h1H2,2H,4H2,5H,(H,6,7)/t2-/m0/s1 c00740= 1.12Beta/C3H7NO3/c4-2(1-5)3(6)7/h1H2,2H,4H2,5H,(H,6,7)/t2-/m1/s1 c00072= 1.12Beta/C6H8O6/c7-1-2(8)5-3(9)4(10)6(11)12-5/h1H2,2H,5H,7-10H/t2-,5+/m0/s1 c06430= 1.12Beta/C6H8O6/c7-1-2(8)5-3(9)4(10)6(11)12-5/h1-5H,8-10H/t2-,3+,4+,5-/m1/s1 c03289= 1.12Beta/C6H8O6/c7-1-2(8)5-3(9)4(10)6(11)12-5/h1H2,2-3H,5H,7-9H/t2-,3+,5+/m0/s1 c01733= 1.12Beta/C5H11NO2S/C1-9-3-2-4(6)5(7)8/h1H3,2-3H2,4H,6H2,(H,7,8) c00855= 1.12Beta/C5H11NO2S/C1-9-3-2-4(6)5(7)8/h1H3,2-3H2,4H,6H2,(H,7,8)/t4-/m1/s1 c00073= 1.12Beta/C5H11NO2S/C1-9-3-2-4(6)5(7)8/h1H3,2-3H2,4H,6H2,(H,7,8)/t4-/m0/s1 c01602= 1.12Beta/C5H12N2O2/c6-3-1-2-4(7)5(8)9/h1-3H2,4H,6-7H2,(H,8,9) c00077= 1.12Beta/C5H12N2O2/c6-3-1-2-4(7)5(8)9/h1-3H2,4H,6-7H2,(H,8,9)/t4-/m1/s1 c00515= 1.12Beta/C5H12N2O2/c6-3-1-2-4(7)5(8)9/h1-3H2,4H,6-7H2,(H,8,9)/t4-/m0/s1 c00806= 1.12Beta/C11H12N2O2/C12-9(11(14)15)5-7-6-13-10-4-2-1-3-8(7)10/h1-4H,5H2,6H,9H,12H2,13H,(H,14,15) c00525= 1.12Beta/C11H12N2O2/C12-9(11(14)15)5-7-6-13-10-4-2-1-3-8(7)10/h1-4H,5H2,6H,9H,12H2,13H,(H,14,15)/t9-/m1/s1 c00078= 1.12Beta/C11H12N2O2/C12-9(11(14)15)5-7-6-13-10-4-2-1-3-8(7)10/h1-4H,5H2,6H,9H,12H2,13H,(H,14,15)/t9-/m0/s1 c00079= 1.12Beta/C9H11NO2/C10-8(9(11)12)6-7-4-2-1-3-5-7/h1-5H,6H2,8H,10H2,(H,11,12)/t8-/m0/s1 c02265= 1.12Beta/C9H11NO2/C10-8(9(11)12)6-7-4-2-1-3-5-7/h1-5H,6H2,8H,10H2,(H,11,12)/t8-/m1/s1 c02057= 1.12Beta/C9H11NO2/C10-8(9(11)12)6-7-4-2-1-3-5-7/h1-5H,6H2,8H,10H2,(H,11,12) c06420= 1.12Beta/C9H11NO3/C10-8(9(12)13)5-6-1-3-7(11)4-2-6/h1-4H,5H2,8H,10H2,11H,(H,12,13)/t8-/m1/s1 c01536= 1.12Beta/C9H11NO3/C10-8(9(12)13)5-6-1-3-7(11)4-2-6/h1-4H,5H2,8H,10H2,11H,(H,12,13) c00082= 1.12Beta/C9H11NO3/C10-8(9(12)13)5-6-1-3-7(11)4-2-6/h1-4H,5H2,8H,10H2,11H,(H,12,13)/t8-/m0/s1 c00083= 1.12Beta/C24H38N7O19P3S/C1-24(2,19(37)22(38)27-4-3-13(32)26-5-6-54-15(35)7-14(33)34)9-47-53(44,45)50-52(42,43)46-8-12-18(49-51(39,40)41)17(36)23(48-12)31-11-30-16-20(25)28-10-29-21(16)31/h1-2H3,3-9H2,10-12H,17-19H,23H,36-37H,(H,26,32)(H,27,38)(H,33,34)(H,42,43)(H,44,45)(H2,25,28,29)(H2,39,40,41)/t12-,17-,18-,19?,23-/m1/s1 c03188= 1.12Beta/C24H38N7O19P3S/C1-24(2,19(37)22(38)27-4-3-13(32)26-5-6-54-15(35)7-14(33)34)9-47-53(44,45)50-52(42,43)46-8-12-18(49-51(39,40)41)17(36)23(48-12)31-11-30-16-20(25)28-10-29-21(16)31/h1-2H3,3-9H2,10-12H,17-19H,23H,36-37H,(H,26,32)(H,27,38)(H,33,34)(H,42,43)(H,44,45)(H2,25,28,29)(H2,39,40,41)/t12-,17-,18-,19?,23-/m1/s1 c00085= 1.12Beta/C6H13O9P/c7-2-6(10)5(9)4(8)3(15-6)1-14-16(11,12)13/h1-2H2,3-5H,7-10H,(H2,11,12,13)/t3-,4-,5+,6?/m1/s1 c06312= 1.12Beta/C6H13O9P/c7-2-6(10)5(9)4(8)3(15-6)1-14-16(11,12)13/h1-2H2,3-5H,7-10H,(H2,11,12,13)/t3-,4+,5+,6?/m0/s1 c05345= 1.12Beta/C6H13O9P/c7-2-6(10)5(9)4(8)3(15-6)1-14-16(11,12)13/h1-2H2,3-5H,7-10H,(H2,11,12,13)/t3-,4-,5+,6-/m1/s1 c01097= 1.12Beta/C6H13O9P/c7-2-6(10)5(9)4(8)3(15-6)1-14-16(11,12)13/h1-2H2,3-5H,7-10H,(H2,11,12,13)/t3-,4+,5+,6?/m1/s1 c00283= 1.12Beta/H2S/h1H2 c00087= 1.12Beta/H2S/h1H2 c00090= 1.12Beta/C6H6O2/c7-5-3-1-2-4-6(5)8/h1-4H,7-8H c05060= 1.12Beta/C6H6O2/c7-5-3-1-2-4-6(5)8/h1-5H,7H c01785= 1.12Beta/C6H6O2/c7-5-3-1-2-4-6(5)8/h1-4H,7-8H c01172= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13)/t2-,3-,4-,5-,6+/m0/s1 c02962= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13) c00275= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13)/t2-,3-,4+,5+,6-/m1/s1 c00668= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13)/t2-,3-,4+,5-,6+/m1/s1 c01113= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13)/t2?,3?,4-,5+,6?/m0/s1 c03735= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13)/t2?,3?,4?,5?,6-/m0/s1 c02965= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13) c00092= 1.12Beta/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h1H2,2-10H,(H2,11,12,13)/t2-,3-,4+,5-,6?/m1/s1 c00623= 1.12Beta/C3H9O6P/c4-1-3(5)2-9-10(6,7)8/h1-2H2,3-5H,(H2,6,7,8)/t3-/m1/s1 c03189= 1.12Beta/C3H9O6P/c4-1-3(5)2-9-10(6,7)8/h1-2H2,3-5H,(H2,6,7,8) c00093= 1.12Beta/C3H9O6P/c4-1-3(5)2-9-10(6,7)8/h1-2H2,3-5H,(H2,6,7,8)/t3-/m0/s1 c00095= 1.12Beta/C6H12O6/c7-1-3-4(9)5(10)6(11,2-8)12-3/h1-2H2,3-5H,7-11H/t3-,4-,5+,6?/m1/s1 c01719= 1.12Beta/C6H12O6/c7-1-3-4(9)5(10)6(11,2-8)12-3/h1-2H2,3-5H,7-11H/t3-,4-,5+,6-/m0/s1 c02336= 1.12Beta/C6H12O6/c7-1-3-4(9)5(10)6(11,2-8)12-3/h1-2H2,3-5H,7-11H/t3-,4-,5+,6-/m1/s1 c01496= 1.12Beta/C6H12O6/c7-1-3-4(9)5(10)6(11,2-8)12-3/h1-2H2,3-5H,7-11H/t3-,4-,5+,6-/m1/s1 c00096= 1.12Beta/C16H25N5O16P2/C17-16-19-12-6(13(28)20-16)18-3-21(12)14-10(26)8(24)5(34-14)2-33-38(29,30)37-39(31,32)36-15-11(27)9(25)7(23)4(1-22)35-15/h1-2H2,3-5H,7-11H,14-15H,22-27H,(H,29,30)(H,31,32)(H3,17,19,20,28)/t4-,5-,7-,8-,9+,10-,11+,14-,15-/m1/s1 c00394= 1.12Beta/C16H25N5O16P2/C17-16-19-12-6(13(28)20-16)18-3-21(12)14-10(26)8(24)5(34-14)2-33-38(29,30)37-39(31,32)36-15-11(27)9(25)7(23)4(1-22)35-15/h1-2H2,3-5H,7-11H,14-15H,22-27H,(H,29,30)(H,31,32)(H3,17,19,20,28)/t4-,5-,7-,8-,9+,10-,11-,14?,15?/m1/s1 c01581= 1.12Beta/C16H25N5O16P2/C17-16-19-12-6(13(28)20-16)18-3-21(12)14-10(26)8(24)5(34-14)2-33-38(29,30)37-39(31,32)36-15-11(27)9(25)7(23)4(1-22)35-15/h1-2H2,3-5H,7-11H,14-15H,22-27H,(H,29,30)(H,31,32)(H3,17,19,20,28)/t4-,5+,7?,8+,9?,10+,11?,14+,15?/m0/s1 c02280= 1.12Beta/C16H25N5O16P2/C17-16-19-12-6(13(28)20-16)18-3-21(12)14-10(26)8(24)5(34-14)2-33-38(29,30)37-39(31,32)36-15-11(27)9(25)7(23)4(1-22)35-15/h1-2H2,3-5H,7-11H,14-15H,22-27H,(H,29,30)(H,31,32)(H3,17,19,20,28)/t4-,5+,7+,8+,9-,10+,11-,14+,15+/m0/s1 c00793= 1.12Beta/C3H7NO2S/c4-2(1-7)3(5)6/h1H2,2H,4H2,7H,(H,5,6)/t2-/m1/s1 c00736= 1.12Beta/C3H7NO2S/c4-2(1-7)3(5)6/h1H2,2H,4H2,7H,(H,5,6) c00097= 1.12Beta/C3H7NO2S/c4-2(1-7)3(5)6/h1H2,2H,4H2,7H,(H,5,6)/t2-/m0/s1 c00100= 1.12Beta/C24H40N7O17P3S/C1-4-15(33)52-8-7-26-14(32)5-6-27-22(36)19(35)24(2,3)10-45-51(42,43)48-50(40,41)44-9-13-18(47-49(37,38)39)17(34)23(46-13)31-12-30-16-20(25)28-11-29-21(16)31/h1-3H3,4-10H2,11-13H,17-19H,23H,34-35H,(H,26,32)(H,27,36)(H,40,41)(H,42,43)(H2,25,28,29)(H2,37,38,39)/t13-,17-,18-,19?,23-/m1/s1 c02843= 1.12Beta/C24H40N7O17P3S/C1-4-15(33)52-8-7-26-14(32)5-6-27-22(36)19(35)24(2,3)10-45-51(42,43)48-50(40,41)44-9-13-18(47-49(37,38)39)17(34)23(46-13)31-12-30-16-20(25)28-11-29-21(16)31/h1-3H3,4-10H2,11-13H,17-19H,23H,34-35H,(H,26,32)(H,27,36)(H,40,41)(H,42,43)(H2,25,28,29)(H2,37,38,39)/t13-,17-,18-,19?,23-/m1/s1 c02187= 1.12Beta/C24H40N7O17P3S/C1-4-15(33)52-8-7-26-14(32)5-6-27-22(36)19(35)24(2,3)10-45-51(42,43)48-50(40,41)44-9-13-18(47-49(37,38)39)17(34)23(46-13)31-12-30-16-20(25)28-11-29-21(16)31/h1-3H3,4-10H2,11-13H,17-19H,23H,34-35H,(H,26,32)(H,27,36)(H,40,41)(H,42,43)(H2,25,28,29)(H2,37,38,39)/t13-,17-,18-,19?,23-/m1/s1
Searching for InChI strings using htDig
htDig is opensource indexing and searching software, having a highly flexible configuration interface. The pertinent extract from the configuration file is shown below:
start_url: http://www.ch.ic.ac.uk/motm/perkin/ \ http://ecrystals.chem.soton.ac.uk/view/ \ http://pubs.acs.org/subscribe/journals/jacsat/suppinfo/ja046734j/ja046734jsi20040621_095820.pdf \ http://wwmm.ch.cam.ac.uk/data/kegg/ http://www.rsc.org/suppdata/OB/b4/b410732b/original/ max_head_length: 10000 maximum_word_length: 255 server_wait_time: 1 max_doc_size: 32000000 #External parsers external_parsers: application/pdf->text/html /var/www/htdig/bin/doc2html/pdf2html.pl \ chemical/x-cml->text/html-internal /var/www/htdig/bin/runtextdig.sh \ image/svg+xml->text/html-internal /var/www/htdig/bin/runtextdig.sh \ chemical/x-mdl-molfile->text/plain-internal /var/www/htdig/bin/runtextdig.sh
This defines the sites to be searched (starting at the top level of each site, and indexing all lower content), various limits, the most important of which is the maximum word length, and External parsers to handle non-standard document types.
This index includes the following sites (with an example from each site shown in brackets)
- http://wwmm.ch.cam.ac.uk/data/kegg/: 1.12Beta/C5H11NO/c6-4-2-1-3-5-7/h1-4H2,5H,6H2
- http://www.ch.ic.ac.uk/motm/perkin/: 1.12Beta/C26H22N4/c1-17-8-10-19(11-9-17)28-20-12-13-23-25(15-20)30(21-6-4-3-5-7-21)26-16-22(27)18(2)14-24(26)29-23/h1-2H3,3-16H,(H2,27,28)/p+1
- http://www.rsc.org/suppdata/OB/b4/b410732b/:
INChI=1.11Beta/C48H37N4O2.Ni/c1-29-5-13-33(14-6-29)45-37-21-22-38(49-37)46(34-15-7-30(2)8-16-34)40-24-26-42(51-40)48(36-19-11-32(4)12-20-36)44(28-54)52-43(27-53)47(41-25-23-39(45)50-41)35-17-9-31(3)10-18-35;/h1-4H3,5-28H,(H-,49,50,51,52,53,54);/q-1;+4/p-1
- http://ecrystals.chem.soton.ac.uk/view/: INChI=1.12Beta/4C11H29Cl2N4O6P3/c4*12-24-15-25(13,17-26(16-24)14-2-1-3-23-26)22-11-9-20-7-5-18-4-6-19-8-10-21-24/h4*1-11H2,14-17H,24-26H
- http://pubs.acs.org/subscribe/journals/jacsat/suppinfo/ja046734j/ja046734jsi20040621_095820.pdf: Trefoilene (sorry, no INChI there yet!)
A typical result (using the default search query) is as follows
ChemDig Search results for:
'112betac26h22n4c11781019 and 11917 and 282012132325 and
1520 and 30 and 216435721 and 261622 and 27 and 18 and 2
and 1424 and 26 and 2923h12h3 and 316h and h2 and 27 and 28
and p and 1' Documents 1 - 7 of 7 matches. More  's indicate a better match.
's indicate a better match.
- [mauveine.xml]









 100
100 - ... ;gt;
</cml:metadataList> <cml:identifier
version="1.12Beta" tautomeric="0">
<cml:basic>1.12Beta/C26H22N4/c1-17-8-10-19(
11-9-17)28-20-12-13-23-25(
15-20)30(21-6-4-3-5-7-21)
26-16-22(27)18(2)
14-24(26)29-23/h1-2H3,
3-16H,(H2,27,28)/p+1</cml
...
File located at: http://www.ch.ic.ac.uk/motm/perkin/mauveine.xml 11/22/04, 3615 bytes. 1.12Beta/C26H22N4/c1-17-8-10-19(11-9-17)28-20-12-13-23-25(15-20 ...
- [mauveine.mol]









 1
1 - mauveine
1.12Beta/C26H22N4/c1-17-8-10-19(11-9-17)
28-20-12-13-23-25(15-20)30(
21-6-4-3-5-7-21)26-16-22(27)18(2)
14-24(26)29-23/h1-2H3,
3-16H,(H2,27,28)/p+1 53 57 0 0 0 0 0 0 0 0 0
v2000 -4.7007 1.2930 -0.0384 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4414 0.1480 -0.0022 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.7771
-1.1443 0.0522...
File located at: http://www.ch.ic.ac.uk/motm/perkin/mauveine.mol 11/22/04, 5183 bytes. Mauveine MDL Molfile
- [Mauveine.rss]









 1
1 - (None of the search words were found in the top of
this document.)
File located at: http://www.ch.ic.ac.uk/motm/perkin/Mauveine.rss 11/23/04, 232838 bytes. Mauveine RSS
- [mauveine.cml]









 1
1 -
1.12Beta/C26H22N4/c1-17-8-10-19(11-9-17)
28-20-12-13-23-25(15-20)30(
21-6-4-3-5-7-21)26-16-22(27)18(2)
14-24(26)29-23/h1-2H3,
3-16H,(H2,27,28)/p+1
File located at: http://www.ch.ic.ac.uk/motm/perkin/mauveine.cml 11/22/04, 3615 bytes. Mauveine CML
- [Mauveine.svg]









 1
1 - (None of the search words were found in the top of
this document.)
File located at: http://www.ch.ic.ac.uk/motm/perkin/Mauveine.svg 11/23/04, 30207 bytes. Mauveine SVG
- [Mauveine.pdf]









 1
1 - ... as a dyestuff. A Postscript to
the postscript To aid molecular discovery, the INChI
identifiers for mauveine and alizarin are included here
as:
1.12Beta/C26H22N4/c1-17-8-10-19(11-9-17)
28-20-12-13-23-25(15-20)30(
21-6-4-3-5-7-21)26-16-22(27)18(2)
14-24(26)29-23/h1-2H3,
3-16H,(H2,27,28)/p+1
1.12Beta/C14H8O7S...
File located at: http://www.ch.ic.ac.uk/motm/perkin/Mauveine.pdf 11/22/04, 161712 bytes. Mauveine Acrobat
- Mauveine









 1
1 - ... as a dyestuff. A Postscript to
the postscript To aid molecular discovery, the INChI
identifiers for mauveine and alizarin are included here
as:
1.12Beta/C26H22N4/c1-17-8-10-19(11-9-17)
28-20-12-13-23-25(15-20)30(
21-6-4-3-5-7-21)26-16-22(27)18(2)
14-24(26)29-23/h1-2H3,
3-16H,(H2,27,28)/p+1
1.12Beta/C14H8O7S...
File located at: http://www.ch.ic.ac.uk/motm/perkin/ 11/23/04, 16204 bytes.
1: http://www.dogpile.com/info.dogpl/search/help/aboutmetasearch.htm
2: http://ecrystals.chem.soton.ac.uk/
3: Algorithm for generation of unique SMILES notation - D Weininger, A Weininger, JL Weininger J. Chem. Inf. Comput. Sci, 1989
* Since these searches were performed (2004-11), GoogleTM has increased the maximum number of tokens used for a query from 10 to 32 (2005-02). Thus the same hits are now not returned for "twas brillig and the slithy toves did gyre and gimble in the wabe" and "twas brillig and the slithy toves did gyre and gimble in the bath". However, "twas brillig, and the slithy toves did gyre and gimble in the wabe all mimsy were the borogoves, and the mome raths outgrabe. Beware the Jabberwock, my son! The jaws that bite, the claws that catch!" and "twas brillig, and the slithy toves did gyre and gimble in the wabe all mimsy were the borogoves, and the mome raths outgrabe. Beware the Jabberwock, my son! The jaws that bite, the rods that fish!" do return the same hits.
Consequently, GoogleTM can now distinguish between InChIs which differ anywhere up to and including the 32nd token. For example at 2004-11 the InChIs below for two enantiomers were seen as identical by GoogleTM whereas as of 2005-02 it can discriminate between them.
c00133= 1.12Beta/C3H7NO2/C1-2(4)3(5)6/h1H3,2H,4H2,(H,5,6)/t2-/m1/s1 c00041= 1.12Beta/C3H7NO2/C1-2(4)3(5)6/h1H3,2H,4H2,(H,5,6)/t2-/m0/s1