Get to know our molecular engineering journal
Crossing the boundary between chemistry and chemical engineering, Molecular Systems Design & Engineering is a hub for cutting-edge research into how understanding of molecular properties, behaviour and interactions can be used to design and assemble better materials, systems and processes to achieve specific functions.
Readers can expect high quality experimental, theoretical and computational research, bringing together physics, biology, chemistry, engineering and materials science.
Authors can expect international exposure, rapid times to publication and efficient peer review. This journal is a partnership between the Royal Society of Chemistry and the Institution of Chemical Engineers (IChemE), which means an expanded community of scientists and engineers will benefit from the articles we publish.
Editorial Board Chair, Juan de Pablo
Institute for Molecular Engineering, University of Chicago, USA
Juan de Pablo is Liew Family Professor in Molecular Engineering at the University of Chicago’s Institute for Molecular Engineering, USA, and Molecular Systems Design & Engineering Editorial Board Chair.

Content highlights
Everything published in 2016 and 2017 is free to read through registered institutions or a free personal account, giving you easy access to the best and latest research, including:
Wenjie Xia, Jake Song, Zhaoxu Meng, Chen Shao and Sinan Keten
Mol. Syst. Des. Eng.., 2016, 1, 40-47
DOI: 10.1039/C6ME00022C
Paper
Enhanced toughness can be achieved through multi-layer graphene (MLG) yielding mechanism in MLG–polymer layered assemblies.
A road towards 25% efficiency and beyond: perovskite tandem solar cells
T. Todorov, O. Gunawan and S. Guha
Mol. Syst. Des. Eng., 2016, 1, 370-376
DOI: 10.1039/C6ME00041J
Review Article
For decades, the appealing potential of tandem solar cells for efficiencies beyond the single-junction Shockley–Queisser limit has led researchers to develop thin film tandem solutions for high performance low cost solar cells.
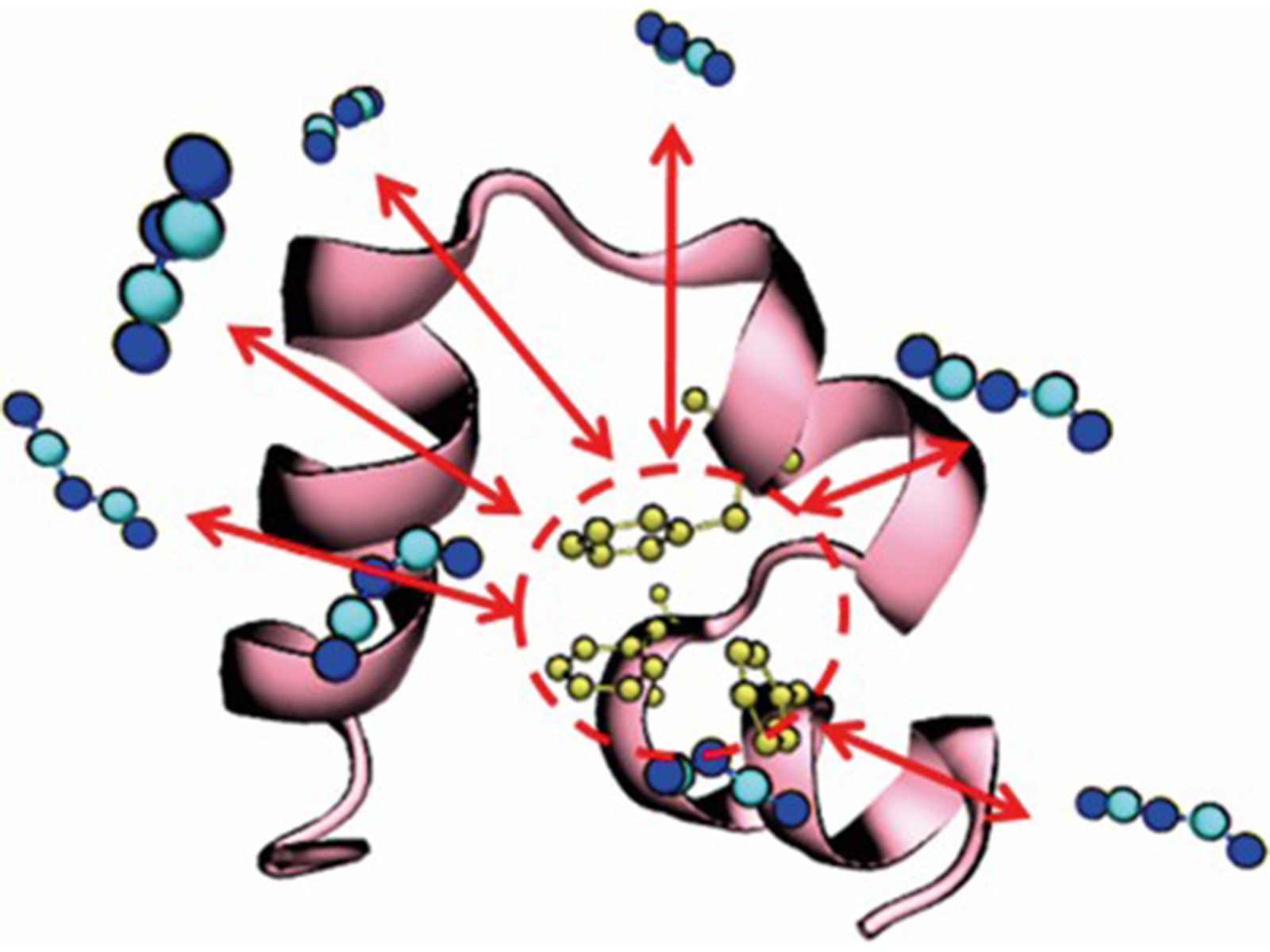
Kinetics and mechanism of ionic-liquid induced protein unfolding: application to the model protein HP35
Hsin-Ju Tung and Jim Pfaendtner
Mol. Syst. Des. Eng., 2016, 1, 382-390
DOI: 10.1039/C6ME00047A
Communication
We demonstrate an approach to quantify protein unfolding times using molecular simulation in a greatly accelerated manner compared to standard MD simulations, showing up to 400 fold speed increases.
Want more from Molecular Systems Design & Engineering?
If you are interested in the work we publish, curious about new developments in our scope or considering the journal for your own work, complete our form now to receive carefully created alerts.
